 Leroy Hood is president and co-founder of the Institute for Systems Biology (ISB). He is also senior vice president and chief science officer of Providence St. Joseph Health. Hood is considered one of the pioneers of systems biology and systems medicine. He is one of only 15 individuals elected to all three US National Academies — the National Academy of Science, the National Academy of Engineering, and the National Academy of Medicine. Hood played a key role in founding or co-founding 15 different biotechnology companies, including Amgen, Applied Biosystems, Rosetta, and Arivale. Hood was the recipient of the 1987 Albert Lasker Basic Medical Research Award for translating the genetics of antibody diversity into explicit molecular terms.
Leroy Hood is president and co-founder of the Institute for Systems Biology (ISB). He is also senior vice president and chief science officer of Providence St. Joseph Health. Hood is considered one of the pioneers of systems biology and systems medicine. He is one of only 15 individuals elected to all three US National Academies — the National Academy of Science, the National Academy of Engineering, and the National Academy of Medicine. Hood played a key role in founding or co-founding 15 different biotechnology companies, including Amgen, Applied Biosystems, Rosetta, and Arivale. Hood was the recipient of the 1987 Albert Lasker Basic Medical Research Award for translating the genetics of antibody diversity into explicit molecular terms.
Q: You received a Lasker Award in 1987 for work that you did in molecular biology towards elucidating the genetic basis of antibody diversity. What prompted you to transition from the field of immunology to developing technologies and improving strategies for quantitative biology?
A: The fact is that there was never really a transition. I’ve been doing biology and technology in parallel for my entire career. Biology always drove the nature of the technology we developed. When I went to Caltech in 1970, I told the chair that I was going to have half my lab doing technology development and half my lab doing molecular immunology. What really moved me was the complexity of biology and disease and the realization that it was necessary to develop a conceptual framework for thinking about this complexity. Later that became systems biology, but at the time we needed to develop a series of tools to let us explore different dimensions of data space. There was a series of paradigm changes that became apparent over time and that were needed to address this complexity. The first paradigm change was bringing engineering to biology. The instruments that my labs developed — the automated DNA and protein sequencers and synthesizers, the ink-jet printer DNA synthesizer, and the Nanostring instruments to carry out single-molecule analyses of mRNA molecules — initiated the idea of high-throughput biological data, and this later led to big data and its analytics.
The Human Genome Project was a second paradigm change. Thanks to the automated DNA sequencer, I was invited to the first meeting ever held on the genome project. Twelve of us met at Santa Cruz in the spring of 1985 to consider whether sequencing the human genome was feasible, and we concluded that it was but were really split on whether it was a good idea. Ninety percent of the biological community was opposed to the genome project in the early days. The NIH [US National Institutes of Health] itself was bitterly opposed to the genome project because it was big science, and big science automatically was bad.
A third paradigm change was creating the first department of cross-disciplinary biology. In 1992, with the help of Bill Gates, I founded the Department of Molecular Biotechnology at the University of Washington, which was enormously successful in tool creation for genomics, proteomics, and computation.
I planned to build in connection with the Department of Molecular Biotechnology a systems biology center but found the University of Washington bureaucracy inhibiting to the development of many aspects of systems biology. So in 2000, Alan Aderem, Ruedi Aebersold, and I co-founded the ISB, and we pioneered systems concepts, applying them to both to biology and medicine. This was the fourth paradigm change.

Institute for Systems Biology
The fifth paradigm change included the concepts of systems medicine and the vision that healthcare should be predictive, preventive, personalized, and participatory — P4 healthcare. At ISB we realized that we needed to catalyze a series of new instruments and technologies to explore different areas of patient data space. We created a strategic partnership at the federal level with the minister of finance for the state of Luxembourg from 2008 to 2013. Over 5 years, Luxembourg gave us $100 million to develop technologies and strategies for systems biology. This led to more than 10 new technology/strategy developments, which placed systems medicine and P4 healthcare at a real tipping point around 2013.
In 2014, we recruited 108 healthy individuals to undergo a yearlong wellness study employing dense, dynamic personal data clouds to discern the key elements of health and wellness. The studies turned out to be absolutely spectacular both with regard to optimizing individual wellness and with regard to the biological secrets the individual data clouds revealed. From that emerged the sixth paradigm change of ‘scientific wellness’, its enormous value to patients, and that idea that wellness, in addition to disease, should be one of the two pillars of healthcare.
In April of 2016, ISB affiliated with the Providence St. Joseph Health systems and became its research arm, with me as the chief scientist. This seventh paradigm change allowed us to start bringing P4 medicine directly to the patient’s bedside.
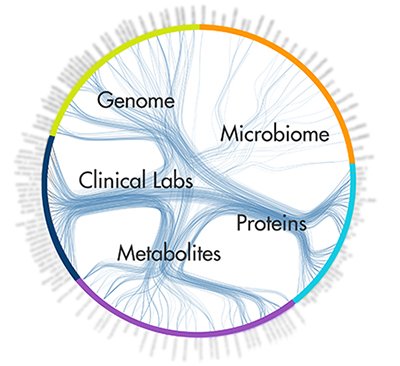
Dense correlations network. Credit: Institute for Systems Biology and Arivale
Q: Can you give a specific example of how scientific wellness and P4 medicine will improve health in a different way than current medical practice?
A: P4 medicine differs from contemporary medicine in that it is proactive and not reactive. It is focused on the individual and not the population. It is all about wellness as well as disease, and not just disease alone. P4 medicine also argues against the way we do current clinical trials with, for example, 20,000 individuals pooled together and given the drug or a placebo. Each individual is unique, with unique genetics and environmental exposure. Individuals shouldn’t be averaged, as that approach just increases the noise over signal for effective drug identification. Drug companies have proven this over and over again. The latest Alzheimer’s drug failure on the part of [Eli] Lilly is a great example. Instead, P4 medicine would create these dense dynamic data clouds for those 20,000 people, and then from each of those data clouds we could stratify the patients according to how well they respond or fail to respond to the drug, or we could delineate whatever features we are interested in. Systems medicine naturally suggests that for many diseases one drug will never be sufficient. We will need three or four drugs and multitherapy approaches.
As an example of how scientific wellness can improve health, we were shocked to learn that 91 out of the 108 pioneers that we studied were low in vitamin D. Of course, vitamin D levels can be increased with supplements, and we thought it would be very easy to bring all those individuals’ levels up to normal. Indeed, we found that giving 1,000 international units (IU) of vitamin D did bring some of them up to normal levels, but many were resistant to increasing their levels at this dosage. Later we discovered that there are six variants in three genes that block the uptake of vitamin D, and according to how many of these blocking variants a person has, you need to prescribe greater doses of vitamin D to change the level. For some people we had to prescribe 15,000–20,000 IU to even get them to budge up towards normal. Putting together the genomic analysis with the clinical chemistry of the level of vitamin D gave us a deep insight that people hadn’t had before.
Credit: Institute for Systems Biology
Q: How would these very large volumes of data collected for each individual be analyzed and made sense of?
A: I am confident that we can handle the computational aspects of data. We extracted an enormous amount of data to analyze from each of the 108 individuals involved in our study, and from these data we have learned several things. First, we have been able to make more than 4,000 fascinating correlations of data bits with one another, and these correlations lead to new approaches to diagnostics and therapeutics, they lead to the ability to think about stratifying a disease into its different subgroups, and they show the interactions of biological systems that we had no idea were interacting. Second, since we had for each individual complete genome sequence data, we showed that we could use GWAS [genome-wide association study] data to calculate the genetic risks in each individual for the 70 or 80 diseases for which there are sufficient GWAS data.
An important point is that some of these genetic risks are actionable, so we can give that information to the patients and have them act on it to avoid disease. For example, we had one participant who had hemochromatosis, which leads to very high iron levels in the blood and ultimately arthritis, beta cell degeneration, liver fibrosis, and cardiac decompensation, which in turn leads to an irreversible chronic disease costing society significantly. You control hemochromatosis by withdrawing a unit of blood every month until you bring the iron level down to normal. This way we reversed an early-stage, rather severe form of hemochromatosis back to normal for this person. His physicians had never been able to figure out the origins of his presenting arthritis. So dynamic data clouds can transform and optimize the wellness of individuals and help avoid or reverse disease.
Q: You mention results from GWAS data analysis that patients could act upon. What about results where it is still unclear what the patient should do to reverse or prevent a disease — how would people deal with that kind of uncertainty?
A: My own feeling is that we should take advantage of all the actionable possibilities. For example, variants in our genomes can tell us how we should diet and exercise; they can let us deal with nutritional insufficiencies and tell us how to deal with inflammation. Sixty-eight percent of the original pioneers had significant inflammation, and most of it was either dietary or genetic deficiencies that could be reversed with supplements. So there are many actionable types of findings.
On the other hand, we can also determine the patient’s genetic risk for Alzheimer’s disease or Crohn’s disease, and these are conditions that we cannot deal with now. What we found in the 108 individuals, after appropriate education about human genetics, is that the majority of them wanted to know about their genetic risk for non-actionable diseases so that they would be warned about the future and could follow the development of new cures and new approaches for their disease risks. Some would prefer not to know that they are at high risk for diseases such as Alzheimer’s, and then I think you don’t give those people this information. It should be the patient’s choice whether they have the knowledge or not. It is really important to educate patients about genetics and what the multi-‘omics’ analyses are, to give them the right framework for thinking about these analytical tools and about both the actionable possibilities and the non-actionable indications so that they can make those choices.
Q: And who should educate the patients?
A: The most effective way to educate the patients is to educate them at the beginning, when they are in K-12 school. Another key point is that we need coaches to explain to each individual patient the actionable possibilities [from test results] and then put them in the context of their own healthcare objectives. Scientific wellness coaching is going to be an enormous opportunity for young men and women in the future.
Q: What needs to change for systems medicine to become reality? What are the biggest obstacles at the moment?
A: The biggest barriers are ignorance on the part of patients and on the part of physicians and healthcare professionals, and one of the big challenges is going to be education. Systems medicine is built on fundamentally different concepts from what 20th-century medicine was (and still is) teaching students. If you really want to have a 21st-century medical school, you should begin with your freshman class. One interesting approach would be to start by taking each freshman MD student through this dense dynamic personalized data cloud analysis the first year, have them generate their own data and learn how to analyze it, and learn what it can and cannot tell them about their own wellness and disease. Then do that for each of the next 3 years, making the integration of systems thinking into clinical practice increasingly sophisticated.
Q: What are the key scientific areas that still need to be developed so that systems medicine can be implemented effectively? Do we need, for example, better disease biomarkers?
A: I think we know how to identify blood biomarkers very well, and we know how to employ genetics to identify the genes that cause simple diseases. But we must determine how we can use genetics and the omics analyses to deal with complex multigenic diseases, such as bipolar disorders, schizophrenia, and autism, more effectively.
We need to develop a series of technologies that are critical. First, we have to be able to take from a single drop of blood all the billions of measurements we make for scientific wellness and make them incredibly inexpensive. The most expensive part of scientific wellness is all the assays, including whole-genome sequencing, and I’d like to see their costs decline by three orders of magnitude over the next 10 years (from $5000 to $5), just as we did with DNA sequencing. The cost of sequencing has come down a millionfold over 15 years or so. Also, imaging at a molecular level in living organisms is going be key to understanding the dynamics of many different diseases and fundamental biological processes.
Q: What is next for you?
 A: This is the most exciting time in my career, and I believe I still have a good 20 years ahead of me. I think we are at an enormous tipping point where there is going to be a fantastic revolution in medicine. The really interesting question is how rapidly that tipping point is going to cause change in how patients are treated. The partnership with Providence St. Joseph — the largest nonprofit healthcare system in the United States — allows us to bring scientific wellness, as well as the systems approaches and technologies, to the patient’s bedside. This partnership has given us access to the healthcare system, and I think within a few years we will apply the systems thinking and the systems technologies to transform major areas of healthcare and medicine. It is an incredibly exciting time for young people who want to combine science and dealing with the human condition. There has never been a better time to consider biology and medicine as transformational professions.
A: This is the most exciting time in my career, and I believe I still have a good 20 years ahead of me. I think we are at an enormous tipping point where there is going to be a fantastic revolution in medicine. The really interesting question is how rapidly that tipping point is going to cause change in how patients are treated. The partnership with Providence St. Joseph — the largest nonprofit healthcare system in the United States — allows us to bring scientific wellness, as well as the systems approaches and technologies, to the patient’s bedside. This partnership has given us access to the healthcare system, and I think within a few years we will apply the systems thinking and the systems technologies to transform major areas of healthcare and medicine. It is an incredibly exciting time for young people who want to combine science and dealing with the human condition. There has never been a better time to consider biology and medicine as transformational professions.
