“Grow faster, you little bugs,” Stanley Cohen recalls begging his bacteria, in jest, because he was so eager to see the results of his experiments. It was a heady time in early 1973, as scientists shuttled DNA back and forth between Cohen’s laboratory at Stanford School of Medicine and Herbert Boyer’s group at the University of California, San Francisco.

Cohen in the lab
[Courtesy of Jose Mercado, Stanford News Service]
Cohen, who trained as a physician, started his research with an interest in infections. The discovery of penicillin in 1928, and other antibiotics subsequently, seemed to herald the end of infectious diseases. “That turned out not to be the case,” says Cohen. “The reason is that bacteria developed resistance to the antibiotics.” Furthermore, bacteria could swap that resistance amongst themselves, sometimes sharing resistance to multiple drugs at the same time.
This resistance was carried on something scientists called the ‘R-factor’, a piece of DNA bacteria could transfer between themselves. Bacteria have one large chromosome, containing most of their genes. But they can also store genes on smaller, circular pieces of DNA called plasmids. R-factors rode on these plasmids. Only a handful of labs were working on plasmids at the time, and in 1968, Cohen set out to understand how R-factor resistance genes were arranged, controlled, and acquired.
First, researchers in his lab tried to move the R-factor plasmids around themselves. Cohen and technician Christine Miller started by purifying the R-factor plasmids. Then, research technician Annie Chang and Leslie Hsu, a medical student working in the lab, succeeded in putting the purified plasmids into new bacteria in 1971.

Cohen with research associate Annie Chang
[Courtesy of Chuck Painter, Stanford News Service]
Chang and Cohen also figured out how to chop up the plasmids, hoping to separate the genes therein, but the process was random and haphazard. Sometimes the fragments would stick back together into a new circular plasmid, but the researchers could not consistently replicate their results. It was like stirring a big pot of broken spaghetti, hoping some strands would stick together in desired combinations. They needed a method to control the subtraction and addition of genes in plasmids.
A Match Made in Paradise
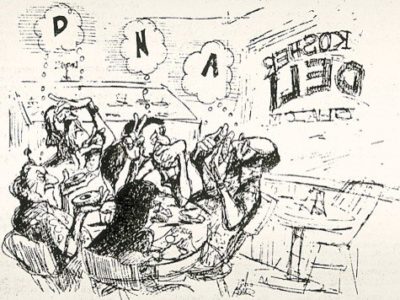
A cartoon depicts the famous Honolulu deli meeting. Stanley Falkow (thinking ”D”), Herbert Boyer (“N”), Stanley Cohen (“A”), with Charles Brinton (lower left) and Ginger Brinton (bottom)
[Courtesy of Dick Adair, Honolulu Advertiser]
Boyer was working on restriction enzymes, protein-based machines that cleave DNA at specific sequences. For example, the enzyme EcoRI cuts only the sequence GAATTC. Those cuts create ‘sticky ends’, so scientists can then glue two EcoRI-cut sequences together in new combinations. Boyer’s group had identified the enzyme in 1968 after isolating it from E. coli cultured from a patient with a drug-resistant urinary tract infection. In Honolulu, Boyer and Cohen plotted their experiments on a napkin.
Back in California, Cohen’s group purified the plasmids from E. coli. Then Chang, who lived in San Francisco, delivered them to Boyer’s lab, where researchers cut the plasmids with restriction enzymes and then glued the pieces together in a different arrangement. They sent the DNA back to Stanford with Chang, where Cohen’s group inserted it into bacteria. Then the scientists isolated the plasmids from those bacteria and sent them back to Boyer for analysis.
Boyer recalled the moment he saw the evidence of rearranged plasmids in a 2009 interview with PLoS Genetics:
“It actually brought tears to my eyes…I knew we’d be able to isolate any piece of DNA that was cut with EcoRI, regardless of where it came from.”
But could they transfer DNA from a different organism into the plasmids carried by E. coli? Many scientists doubted it, assuming that the DNA would be incompatible.
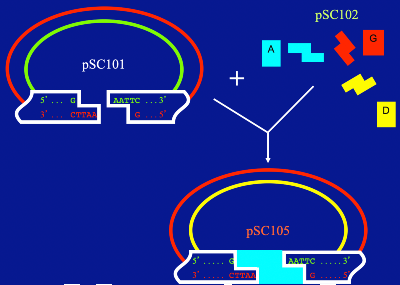
Cutting Cohen’s plasmid, pSC101, with the restriction enzyme EcoRI leaves ”sticky ends.” Other pieces of DNA, cut with the same enzyme, can fit into the sticky ends to create a new plasmid, pSC105.
Nonetheless, Cohen and Chang managed to insert a piece of DNA from the plasmid of another bacterium, Staphylococcus aureus, into E. coli via a plasmid. Then they rejoined forces with Boyer, and in 1973 the collaborators took a piece of DNA from the African clawed frog, a common lab animal, and put that into E. coli with a plasmid too. This linking of disparate DNA sequences would quickly come to be called ‘recombinant DNA technology.’
The Rise of Recombinant DNA Technology
These studies showed that scientists could use plasmids and restriction enzymes to ‘clone’ DNA sequences from any creature. They could make large amounts of the plasmid for further study. Other scientists were excited and requested that Cohen send them his plasmid for their own experiments.
But researchers were worried, too. This new approach also meant that scientists could conceivably create potentially hazardous ‘Frankenorganisms’ expressing unnatural and potentially hazardous genes. What if they transferred antibiotic resistance into bacteria that didn’t already have it, creating new superbugs? Or what if they put cancer-causing viral genes into bacteria and created novel cancer agents?
A committee that included Cohen and Boyer, as well as Paul Berg, evaluated the possible dangers of mixed-DNA organisms. In a 1974 article, the group proposed that scientists should not insert antibiotic resistance genes into species of bacteria that weren’t already known to harbor such resistance, nor should they transfer animal viral genes into plasmids. The National Institutes of Health also issued guidelines, requiring that the riskiest experiments be conducted in airlock-secured facilities, the same kind used to study the Ebola virus and other very-high-risk pathogens.
In a 1977 article in the journal Science, Cohen argued that the risks of genetic engineering were mostly speculative, whereas the benefits were much more likely.
“It really was an experience in trying to educate the political community scientifically,” he recalls. “I did spend a lot of time working with congressmen and senators and their staffs.”
Scientists followed the guidelines, and with time it became clear that Cohen was right about the positive potential of DNA recombination.
Electron micrograph of plasmid DNA
In 1976, Boyer made an initial investment of $500 to co-found Genentech, a pharmaceutical company in Oceanside, California. Genentech scientists copied the human insulin gene into a plasmid and put that into bacteria. The bacteria then used that gene to pump out insulin for people with diabetes. Next, Genentech made human growth hormone, which is used to treat short stature and other related conditions in the same way.
The recombinant DNA revolution had begun. Today, hundreds of biotechnology companies use recombinant DNA technology to make medications for conditions ranging from cancer to hemophilia. In the lab, restriction enzymes and plasmids have become standard tools, enabling scientists to study genes in detail. Scientists transfer new genes into crop plants to make foods more appealing or nutritious. The list goes on.
“You couldn’t do any of the things that people are doing now if you didn’t have the cloning technology and the plasmids,” says Miller, who was a research associate in Cohen’s lab until her retirement in 2011.
Meanwhile, Cohen is still working on the problem of antibiotic resistance, studying how bacteria adapt to stressful conditions. He’s also investigating how repetitive DNA sequences contribute to diseases such as Huntington’s disease and muscular dystrophy.
“He’s just motivated to learn things, to find things out,” says Miller. “He’s still working as hard as ever.”
