
James Rothman
Sloan-Kettering Institute

Randy Schekman
University of California, Berkeley
For discoveries revealing the universal molecular machinery that orchestrates the budding and fusion of membrane vesicles — a process essential to organelle formation, nutrient uptake, and secretion of hormones and neurotransmitters.
This year's Lasker Basic Medical Research Award honors two scientists who discovered the universal molecular machinery that orchestrates the budding and fusion of membrane vesicles, a process that cells use to organize their activities and avert the biochemical anarchy that would result if all of their contents commingled. By 1970, George Palade had published his classic work showing that proteins travel between cellular compartments, but the molecular basis for this phenomenon was unknown. Using a biochemical and genetic approach, respectively, James Rothman and Randy Schekman transformed this descriptive field into one of detailed molecular clarity.
The cellular trafficking system they elucidated underlies numerous vital processes. Their discoveries explain, for example, how pancreatic cells release insulin, how nerve cells communicate, how embryos liberate growth factors to stimulate organ development, and how viruses infect. Alterations in these pathways explain a plethora of pathological processes, including the most common form of diabetes and the lethal effects of bacterial diseases such as botulism and tetanus. Scientists have developed drugs that target the trafficking system and are pursuing additional ways to pharmacologically manipulate it in an attempt to control an increasing number of illnesses, particularly those that relate to brain chemistry.
Although this general picture was clear by the mid-1970s, the details remained obscure. No one understood, for example, how cells made vesicles from membranes, how vesicles knew where to go, or how they fused with their target membranes.
Captivated by these questions, Rothman and Schekman embarked on their scientific quests. Both of them aimed to track down the molecular machinery that performed the crucial trafficking functions, and then to figure out how it worked, but they chose different initial strategies. Rothman decided to dismantle the pathway and reconstitute it from its components in a test tube; success at building up the entire process from its parts would indicate that he had fished out the vital elements. Similar so-called biochemical strategies had boasted victories in the arena of complicated, multi-step biological activities, including the mechanism of DNA replication and protein synthesis. Schekman selected a genetic scheme, in which researchers prod cells to reveal constituents that participate in a particular pathway. The aim was to obtain mutants that can't perform a certain function; the genes mutated in these cells presumably play essential roles in the process of interest. This strategy, too, had triumphed — in elucidating the mechanism by which cells divide, for example. Both approaches provide a way to find single components of complex systems, which can then be probed further.
Each tactic produced significant contributions of its own, and reinforced findings from the other. Together, the advances unmasked the machinery and means by which cells transport proteins, yielding unanticipated insight into fields that span the physiological gamut.
Brave new worlds
When Rothman and Schekman began their work in the late 1970s, some scientists raised their eyebrows; others yawned. Schekman, a biochemist and microscopist, had cooked up the idea of using yeast genetics to identify cellular trafficking components. Grant reviewers trounced his first proposal. He had no experience with yeast, they thought that he underestimated the difficulty of working with membranes, and doubted whether yeast had a robust secretion system anyway. Furthermore, people were skeptical about whether findings from this single-celled creature would apply to animals. Rothman encountered skepticism as well. Most scientists thought his approach would fail because compartments that exchange delivery vesicles in the cell tend to lie close together, and they believed that vesicles found their targets in large part due to this proximity. Breaking cells open would destroy spatial relationships, and the appropriate membranes would never locate each other, the argument went. No one yet thought that specific features inherent to the membranes would direct their movement. Despite these gloomy forecasts, the researchers persevered.
Assuming that yeast would need its vesicle transport system to survive, Schekman reasoned that he wouldn't obtain live mutants whose defects lay in crucial trafficking genes. Instead, he and his graduate student, Peter Novick, sought strains carrying so-called temperature-sensitive mutations. At low temperatures, these defects are mild enough to allow the proteins in which they reside to limp along and function adequately; raising the temperature, however, exacerbates their effects, inactivates the protein, and kills the organism. During its demise, the yeast would exhibit a measurable aberration in the transport process, Schekman reasoned.
Novick and Schekman sifted through 87 temperature-sensitive mutants and found two in which enzymes that normally reside in the cell's outer wall remain stuck inside. Presumably an intracellular traffic jam had detained them and they couldn't escape the cell. Using electron microscopy, the team showed that the cells looked like they had measles; they contained an enormous number of vesicles that could not be discharged.
This strategy produced the first secretory genes, which Schekman named sec1 and sec2. The approach, however, was laborious, and he wanted a quick way to enrich for yeast that did not export proteins properly. Such cells would be denser than normal, he figured. With this rationale, he used centrifugation to collect new candidates from a different mutant collection. Because yeast with secretion defects were heavier than normal, they sank to the tube's bottom, whereas healthy yeast or those with abnormalities in different pathways remained afloat. Schekman then sorted the dense individuals, seeking ones that retained enzymes that they'd normally secrete. This method produced 23 genes involved in secretion, including the first two.
Using electron microscopy, Schekman found that these mutants accrued perturbations in the transport pathway. Some, for example, accumulated vesicles that presumably failed to fuse with their target membranes. Using standard genetic techniques, he figured out the order in which the sec genes acted. Schekman had thus achieved a major victory: He afforded the first peek into intracellular trafficking on a molecular level by identifying genes that promote specific steps required for secretion.
Simultaneously, Rothman had been devising an ingenious test tube system. He exploited the observation that proteins traveling to the cell surface acquire chemical modifications of their sugar side chains at specific points of their journey. One of these structural changes, acquired in the Golgi, renders the side chain resistant to a certain enzyme that Rothman could add to the reaction mixture. Proteins at earlier stages would remain susceptible to cleavage, whereas those that had reached this destination would ignore the enzyme.
To obtain an extract containing a protein he could track, Rothman and his first postdoctoral fellow, Erik Fries, crushed a special line of mammalian cells carrying a newly made radioactive protein destined for the cell surface. These cells also contained a defective version of the protein that normally makes the structural change in sugar side chains. Therefore, even if the radioactive protein in this first extract made it to the Golgi site of interest, it would remain sensitive to the added enzyme. The team then supplemented this first extract with a second extract from a normal cell. Only radioactive protein from the first extract that traveled to the Golgi in the second extract, which harbored a normal version of the protein that modifies the side chain, would resist the added enzyme, the researchers reasoned. The system worked: After combining the contents of the two cells, Rothman detected radioactive protein that the enzyme could not cut. Electron microscopy confirmed these findings. Using extracts from cells with Golgi of different sizes, he established that the radioactive protein had moved from its original compartment in the first extract to the second extract's Golgi. Because the set-up demanded that transport occur between compartments from different cells, the achievement meant that spatial relationships in the cell were not crucial for transport. Rothman had devised a powerful system with which to tease apart the biochemical underpinnings of intracellular trafficking.
Budding insights
To peer more closely at his extracts, Rothman hooked up with Lelio Orci, an accomplished electron microscopist at the University of Geneva. Together, they showed that a molecular coat enrobed — but only where vesicles were budding. Furthermore, these vesicles emerged from one Golgi compartment, lost their coats, and then fused with another. Because Rothman suspected that this coat played an essential role in the process, he wanted to isolate it and identify its subunits. He failed to obtain large enough quantities for analysis, however, because the coat fell off soon after budding.
By this time, Schekman's former student Novick had set up his own lab at Yale and discovered that transport requires a molecule called GTP to activate a protein that acts at a late step in the protein export process. This observation spurred Rothman to ponder possible involvement of GTP in early steps as well. He added a chemical that resembles GTP but that fastens to proteins much more avidly than GTP does, and does not allow them to break it down to a related compound called GDP. This chemical inhibitor blocked transport from the Golgi in Rothman's extracts, suggesting that the reaction employs a protein that normally converts GTP to GDP. Furthermore, the extracts accumulated large numbers of vesicles, as if they could bud properly, but not fuse with their target membranes. The chemical inhibitor thus provided a means to produce copious numbers of vesicles, from which Rothman could harvest and analyze coats.
Seven proteins assembled to compose the coat components. As the inhibitor study had hinted, a GTP-binding protein was required for budding, one that had previously been identified, but whose function had been unknown. When bound to GTP, the protein extends a fatty arm, which seeks to flee the watery environment of the cell's cytosol and insert itself into a greasy membrane. When bound to GDP, the protein tucks away this oily protuberance.
This GTP-GDP switch provided the underpinnings for coat formation — and for budding. In its GTP form, the protein attaches to a membrane and recruits the seven-protein aggregates. These molecular subunits naturally congregate into a curved structure, and Rothman proposed that this behavior helps mold the bud and eventually forces the membrane to pinch off. In this scheme, coat assembly would provide the mechanical force necessary to drive budding. After vesicle formation, the GTP-binding protein converts its GTP to GDP, its blubbery anchor retracts, and the protein disengages from the membrane, taking the coat with it. This scenario explained the inhibitor's effects: Unable to transform GTP to GDP, the protein remained affixed to the membrane and the vesicles could not disrobe. As a result, they could not fuse with their target membranes and accumulated in the extract.
In the meantime, Schekman, having identified a collection of secretory genes, had desired to find out exactly how each one worked. He, too, had concocted a system for studying protein transport — but from yeast. He decided to focus on the ER-to-Golgi transport step. Working with Orci, he showed that this process yielded coated vesicles that looked similar to those that Rothman had seen. Purification of the subunits revealed that the coat consisted of different building blocks from the ones Rothman had identified in his earlier studies of Golgi-to-Golgi transport. Striking similarities emerged, however: A GTP-binding protein moored the coat to the membrane, and later promoted its disassembly after turning its bound GTP into GDP.
Rothman and Schekman had proven that budding requires an ER or a Golgi membrane, coat components, and a GTP-binding protein. Because the same basic mechanism operated at two sites, Schekman wondered whether an artificial membrane consisting of pure lipids would support the budding reaction — and it did. This experiment established that the coat proteins themselves can deform membrane and pinch off vesicles.
EnSNAREd by fusion
Budding explained how protein-laden vesicles leave intracellular pouches, but not how they enter. Before Rothman and Schekman had tied up the mechanism of budding, they had made major progress toward understanding fusion.
Rothman discovered that a compound called N-ethylmaleimide (NEM) blocked trafficking in his test tube system. By separating untreated extracts into portions and identifying which ones rescued the NEM-poisoned reaction, he eventually purified the so-called NEM-sensitive factor (NSF). Another collaboration with Orci revealed that NEM-exposed extracts accumulated vesicles. In contrast to those amassed in extracts treated with the GTP-related compound, these vesicles were naked. Realizing that they had lost their coats, Rothman reasoned that NSF must be important for the step that followed uncoating: fusion.
He tracked down the gene that encoded NSF and sequenced it. It strongly resembled SEC18, a gene that Schekman had isolated in his early experiments. This breakthrough, in 1989, was the first golden spike that linked the two tracks, and convinced many skeptics that both strategies were generating valuable knowledge. The work established that a similar fusion step occurred in distantly related creatures and suggested that NSF/Sec18p was required for multiple fusion events: Golgi-Golgi (from Rothman's work on mammals) and ER-Golgi (from Schekman's work on yeast). The convergence of the two lines of inquiry also established the physiological relevance of Rothman's test tube system and the pertinence of yeast secretion to mammals.
Although fusion needs NSF, the protein cannot promote that reaction on its own. NSF bound to target membranes, but only in the presence of another cytosolic protein, which Rothman called "soluble NSF attachment protein" (SNAP). The SNAP gene turned out to resemble another one of Schekman's genes: SEC17. Simultaneously, Schekman established that the yeast versions of NSF and SNAP collaborate to foster vesicle fusion in living yeast. These findings further corroborated the results from the two systems and bolstered their physiological relevance and generality among distantly related organisms.
Next, Rothman dug deeper into how SNAP and NSF work. SNAP binds to membranes and then NSF attaches to SNAP, he discovered. Unlike the proteins that trigger coat formation, SNAP does not contain a greasy appendage that could tether it to the membrane. Rothman therefore proposed that it needs to hook up with a receptor that resides in the membrane, which he called the SNAP receptor, or SNARE.
Wanting to identify membrane proteins that compose this presumptive SNARE, he and his postdoctoral fellow, Thomas Söllner, ground up brain and sought components that stuck to NSF and SNAP. They had chosen brain because, of the tissues they tested, it harbored the largest concentration of NSF- and SNAP-binding material. Fortuitously, this decision gave rise to insights into neuron signaling as well as fundamentals of membrane trafficking.
Rothman and Söllner retrieved three membrane proteins, which clasped each other tightly. Scientists had previously noticed these molecules at sites in the brain where nerves fire messages, but no one had known their function. One resides in vesicles that carry neurotransmitters; the other two dwell on the cell membrane with which those vesicles fuse. The results immediately implicated these proteins — now called SNAREs — in membrane fusion, and the physical association between them suggested how they might expedite this process.
Rothman conjectured that the SNAREs he discovered united the membranes so that fusion could occur. He then generalized this idea, speculating that all vesicle and target membranes carry proteins (called v- and t-SNAREs, respectively) that recognize each other. If only particular combinations gripped each other, this pairing scheme could explain how certain vesicles docked with certain target membranes. The idea thus encompassed fusion specificity as well as its basic mechanism.
To test his hypothesis that SNARE pairings contain enough information to direct specificity for membrane fusion events, Rothman used the complete yeast genomic sequence to identify the organism's potential SNAREs, some of which Schekman and his coworkers had previously assigned to particular transport steps. He then manufactured all of the v-SNAREs and most of the cell's t-SNAREs, incorporated them into artificial membranes, and assessed which combinations caused membranes to fuse. Only 9 out of 275 combinations passed this test, and all now correspond to known transport pathways. Isolated SNARE proteins can thus reproduce the pattern of membrane flow in the cell. The location of SNARE partners among cellular membranes therefore governs fusion events within the cell.
On his way to this elegant conclusion, Rothman showed that the two SNARE partners on their own promote fusion of artificial membranes. That experiment established that they alone can foster the reaction, an observation that raises the question of what SNAP and NSF do. In independent experiments, William Wickner, of Dartmouth Medical School, showed that these proteins, which had led Rothman to the SNAREs, were required for fusion not because they promote membrane melding, but because they tear apart the two halves of the SNARE couple. Without SNAP and NSF, fusion winds down because the SNARE partners are stuck in a terminal embrace. Axel Brunger, of Yale University, and Reinhard Jahn, of the Max Planck Institute in Göttingen, have elucidated the structural basis for this interaction at the atomic level. The t-SNARE and v-SNARE components wrap around each other, pulling together the membranes in which they're embedded, thus prompting fusion.
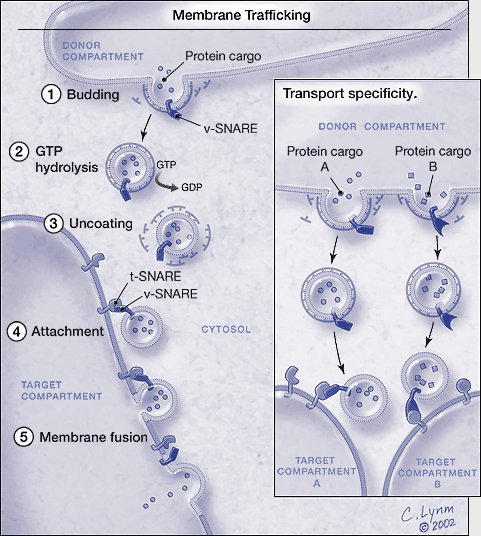
Membrane trafficking. *For a full description of the figure, please see legend at the end of the essay. The graphic is © Cassio Lynm, 2002. To obtain permission for use please contact Cassio Lynm at cassio-lynm.com
Trafficking directed
The combined work of Schekman and Rothman has unveiled the key machinery and mechanisms of membrane trafficking. This process is vital for countless physiological events, and when it malfunctions, illness occurs. In type II diabetes, for example, cells can't ingest glucose — even in the presence of adequate amounts of insulin — because molecules that normally reside on the cell surface and import the sugar are stranded in vesicles inside. Some patients are already using insulin-sensitizing drugs that restore fusion so the transport molecules can reach the surface and slurp the glucose that's just out of reach.
Knowing the mechanism of protein secretion in yeast has allowed scientists to use these microbes as protein factories. Yeast genetically engineered to secrete human insulin currently produce about 25 percent of the world's insulin supply and all of the hepatitis B antigen used for vaccination. Medical applications based on new information about trafficking extend much further. Drugs that shift the balance between neurotransmitter uptake and export might someday provide novel therapies for cognitive and mood disorders. Recent studies suggest, for instance, that regulation of membrane fusion is altered in schizophrenia.
Rothman's and Schekman's legacy has permeated a vast number of physiological disciplines, including neurobiology, endocrinology, virology, and embryology, and promises to expand its impact further. Happily, the doomed projects that began 25 years ago have defied their initial prognoses.
by Evelyn Strauss
*Figure. Membrane trafficking.Vesicles carrying particular proteins bud from the donor compartment, driven by coat assembly. Conversion of guanosine triphosphate (GTP) to guanosine diphosphate (GDP) triggers uncoating. The naked vesicle is then poised to attach to the target compartment through the specific binding between the v-SNAREs and t-SNAREs. The vesicle and the target membranes fuse and the protein cargo arrives at its destination. Only the basic process is shown; genetic and biochemical studies have revealed other proteins that regulate the SNARE-dependent fusion process. Inset: Transport specificity. Particular v- and t-SNAREs mark vesicles and target compartments. Only matched pairs unite and promote fusion. By this method, different intracellular sites obtain and maintain their unique protein constituents. The graphic is © Cassio Lynm, 2002. To obtain permission for use please contact Cassio Lynm at www.cassio-lynm.com.
Key publications of James Rothman
Fries, E. and Rothman, J.E. (1980). Transport of vesicular stomatitis virus glycoprotein in a cell-free extract. Proc. Natl. Acad. Sci. USA. 77, 3870–3874.
Block, M.R., Glick, B.S., Wilcox, C.A., Wieland, F.T. and Rothman, J.E. (1988). Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc. Natl. Acad. Sci. USA. 85, 7852–7856.
Serafini, T., Orci, L., Amherdt, M., Brunner, M., Kahn, R.A., and Rothman, J.E. (1991). ADP-ribosylation factor (ARF) is a subunit of the coat of golgi-derived COP-coated vesicles: A novel role for a GTP-binding protein. Cell. 67, 239–253.
Söllner, T., Whiteheart, S.W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P. and Rothman, J.E. (1993). SNAREs, SNAP receptors implicated in vesicle targeting and fusion. Nature. 362, 318–324.
Weber, T., Zemelman, B.V., McNew, J.A., Westermann, B., Gmachi, M., Parlati, F., Söllner, T.H., and Rothman, J.E. (1998). SNAREpins: Minimal machinery for membrane fusion. Cell. 92, 759–772.
McNew, J.A., Fukuda, R., Parlati, F., Johnston, R.J., Paz, K., Paumet, F., Söllner, T.H., and Rothman, J.E. (2000). Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 407, 152–159.
Key publications of Randy Schekman
Novick, P. and Schekman, R. (1979). Secretion and cell surface growth are blocked in a temperature sensitive mutant of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 76, 1858–1862.
Novick, P., Field, C., and Schekman, R. (1980). The identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 21, 205–215.
Deshaies, R.J. and Schekman, R. (1987). A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 105, 633–645.
Baker, D., Hicke, L., Rexach, M., Schleyer, M., and Schekman, R. (1988). Reconstitution of Sec gene product-dependent intercompartmental protein transport. Cell. 54, 335–344.
Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., Rexach, M., Ravazzola, M., Amherdt, M., and Schekman, R. (1994). COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 77, 895–907.
Matsuoka, K., Orci, L., Amherdt, M., Bednarek, S.Y., Hamamoto, S., Schekman, R., and Yeung, T. (1998). COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 93, 263–275.
Award presentation by Michael Brown
 Today, we honor Jim Rothman and Randy Schekman for unmasking the traffic signals that direct the assembly line of the cell. Each cell is a factory that manufactures thousands of proteins every minute and exports them to the outside world. These proteins are assembled in steps that must be performed in the proper order. To organize this assembly, cells use workstations specialized for each chemical reaction. This solves one problem but it creates another: how to transport partially finished proteins from one workstation to another, and how to make certain that each workstation has the right amount of energy, supplies, and worker enzymes.
Today, we honor Jim Rothman and Randy Schekman for unmasking the traffic signals that direct the assembly line of the cell. Each cell is a factory that manufactures thousands of proteins every minute and exports them to the outside world. These proteins are assembled in steps that must be performed in the proper order. To organize this assembly, cells use workstations specialized for each chemical reaction. This solves one problem but it creates another: how to transport partially finished proteins from one workstation to another, and how to make certain that each workstation has the right amount of energy, supplies, and worker enzymes.
The work stations are called organelles. They are like balloons filled with enzymes enclosed by a membrane. If you were reduced to microscopic size and were standing inside a human cell, you would be surrounded by hundreds of these organelles floating in space. Every so often, you would see a small bubble form on the surface of one organelle. The bubble is called a vesicle and its formation is called budding. The budding vesicle is filled with partially assembled proteins that must be carried to the next organelle. The vesicle detaches from its mother organelle and moves to a target organelle where it sticks like Velcro. Next the membranes between the vesicle and the organelle dissolve and the contents of the vesicle are injected into the organelle. This is called membrane fusion.
In the 1970s, when Rothman and Schekman began their work, most scientists despaired of solving these problems. The issues were too complex for classic biochemistry. The classic biochemist would crack open the cells, isolate vesicles, incubate them in test tubes, and measure the budding and fusion. The responsible proteins would be fractionated and the components would be isolated by biochemical complementation. This reductionist approach was used by Arthur Kornberg, when he made genes in a test tube. But even Kornberg would not dare attack a problem as complex as vesicle budding and fusion. This process must depend on the geometry within the cell. Vesicles bud and fuse at defined positions. Crack open the cell and you lose the geometry.
Up stepped Jim Rothman, a brand new biochemist at Stanford. Jim had the three ingredients essential for scientific success: training, environment, and youth. First: training. Rothman was suckled on membranes — as an undergraduate at Yale, as a graduate student at Harvard, and as a postdoctoral fellow at MIT. Second: environment. Rothman joined Kornberg's department which was built on the premise that no problem in biology is too complex for biochemistry. Third: youth. Rothman was young and fearless — what's more he had an idea.
A biochemist lives and dies by his assay. Rothman thought of a clever assay to measure vesicle budding and fusion in a test tube. His assay overcame the biggest problem — specificity — which arises because membranes are fragile. If you treat them too roughly, the organelles will burst and fuse at random, creating chaos that is impossible to sort out from the true cellular event.
Rothman's assay used mutant animal cells that were generated by Stuart Kornfeld at Washington University. These cells are missing an enzyme that is necessary to complete the assembly of secreted proteins. The proteins accumulate within organelles in a partially finished form. From these cells Rothman obtained organelles that contained partially finished proteins. He mixed these organelles with organelles from normal cells in a test tube. If vesicle budding and fusion occurred, then the unfinished proteins would move to the normal organelles and their assembly would be completed. And that is what Rothman observed. Directional movement in a test tube. The assay wasn't robust, but it allowed the purification of the first enzyme that was required for the fusion process. Like many revolutionary advances, Rothman's work was met with skepticism. Most cell biologists clung to the notion of vitalism — true vesicle movement could occur only in living cells. Rothman dealt a death blow to vitalism.
Before I go to the next chapter, I must introduce Jim's alter ego, Randy Schekman, who also had the three attributes of success. First: training. Randy earned his PhD with Arthur Kornberg and we've already heard what he stands for. He then went to San Diego where he studied with Jonathan Singer, a master of membranes. Second: environment. Randy began his career at Berkeley, where biochemistry was dominated by Daniel Koshland, Jr., himself a Lasker awardee, and a firm believer in the power of biochemistry to unlock all mysteries. Third: youth. Schekman was young, and like Rothman he had an idea and the courage to try it out.
Schekman, like Rothman, was captivated by vesicle budding, targeting, and fusion. So far these two sound like twins. Both were trained in membrane chemistry, both were exposed to Kornberg and both began their careers as Bay Area biochemists in the late 1970s. Both also collaborated with Lelio Orci, the great microscopist of Geneva. But there was an important difference. Instead of tackling this problem through biochemistry, Schekman decided to use genetics. He chose a simple organism, a yeast. The biochemist works with broken cells in test tubes. The yeast geneticist works with living cells in Petri dishes. The geneticist's approach is more conservative because it does not deny vitalism. But Schekman's approach was bold in another way. It assumed that vesicle budding and fusion in a lowly yeast would resemble the process in human cells. After all, we don't study yeast because we want to make better beer or bread. We study yeast to learn something about ourselves. When Schekman started, there was little basis to assume that the process in yeast would be anything like the process in animal cells. The organelles hadn't been described microscopically in yeast. Just as Rothman proceeded on the world view of Kornberg and Koshland, Schekman followed the precedent of Lee Hartwell, another Lasker awardee, who used yeast genetics to dissect the cell cycle and found principles that apply to all cells, including humans.
Like the biochemist, the geneticist needs an assay. Schekman conceived a brilliant and simple one. The challenge was to isolate mutant cells with defects in vesicle budding and fusion. This would unmask the crucial genes. Schekman realized that defects in budding or fusion would jam the cell's assembly line. The exported proteins would accumulate as unfinished goods. This buildup would make the cell heavier. Mutant cells could be separated from wild-type cells by centrifugation. Using this simple trick, Schekman and his students identified more than 70 genes required for vesicular traffic in yeast. These genes encoded proteins. Using classic genetic complementation, they determined the order of each protein in the yeast assembly line.
By 1980, Rothman and Schekman had both established their systems, and now the game began. For the next 20 years, the two played leapfrog. First one would discover a protein and then the other would use that information to discover the next protein. Each would vault to the next breakthrough on the haunches of the other. In a moment you will meet these men and you will note that one has the tougher task in leapfrog.
I don't have time to outline the individual accomplishments of Jim and Randy. They are monumental. Let me summarize by saying that we now have a clear understanding of the mechanisms by which protein cargo is sorted into vesicles, by which vesicles bud from organelles, by which they choose their proper target organelles and fuse with target membranes. These mechanisms are so fundamental that they are conserved from yeast to humans. This conservation allowed yeast genetics to play leapfrog with mammalian biochemistry.
The implications of this work are broad and deep. It gives us the instruction book for the assembly of a cell. It tells us how organelles are organized. The work teaches us how cells manufacture and secrete proteins like insulin and growth hormone that govern our metabolism. How brain cells discharge the chemical transmitters that mediate thought, feeling, and movement. How genetic defects in protein assembly cause human diseases like cystic fibrosis and heart attacks due to hypercholesterolemia. Indeed, we can no longer conceive of any cell, normal or diseased, without thinking of the fundamental processes that were discovered by Jim Rothman and Randy Schekman.
Let me close with an anecdote that shows how genes travel. First, I must tell you that Jim is a fiery and flamboyant orator who loves to challenge his audiences with audacious ideas. Randy is exactly the opposite. He is reserved and conservative, even fastidious. Both Jim and Randy had grandfathers who lived in neighboring villages or shtetls in Bessarabia in Eastern Europe. Rothman's grandfather was the village rabbi, a flamboyant orator, and Schekman's grandfather was the tailor. Genes cross oceans.


