Every year, about 1 in 800 babies in the US is born with Down syndrome. This condition arises from an extra copy of chromosome 21, which is reflected in its other name, trisomy 21. Tests such as amniocentesis and chorionic villus sampling can accurately determine whether a fetus carries such serious chromosomal abnormalities, but these methods require invasive sampling of fetal tissue, so they come with a small but real risk of miscarriage. Because clinicians would like to offer the procedures only to women whose chances of having a child with Down syndrome exceed those of experiencing a miscarriage from amniocentesis or chorionic villus sampling, they try to identify women whose risk of having a sick infant is especially high.
The odds of giving birth to a baby with Down syndrome increase exponentially after age 30: At age 25, it is 1/1340; at age 35, it is 1/350; and at age 40, it is 1/85. Certain biochemical and sonogram findings also reflect risk, and combining these factors provides a screening tool that points toward women who would most benefit from the diagnostic tests. Although such protocols filter out women who are least likely to be carrying babies with Down syndrome, many with healthy pregnancies score high enough that they needlessly face the invasive procedures. These ambiguities have been largely conquered with a simple blood test.
A welcome surprise in the bloodstream
Beginning in the late 1960s, scientists dreamed of obtaining fetal cells from pregnant women's blood, thus affording a glimpse into the fetal genetic makeup without perturbing the pregnancy. Starting when Lo was a medical student in the 1980s, he shared this fantasy. Despite ingenious schemes from numerous laboratories, results continued to disappoint.
In 1996, two reports caught Lo's eye.
One study found tumor DNA in plasma, the liquid, cell-free component of blood; the other found it in serum, which is plasma minus its clotting factors. A fetus shares similarities with tumors, Lo reasoned, as both tissues differ immunologically from cells in the rest of the body and establish a vascular interface with their hosts. If cancer can release genetic material into the cell-free portion of blood, Lo thought, surely a fetus can too.
To probe this idea, Lo extracted DNA from plasma and serum. He then used polymerase chain reaction (PCR), which copies and thus amplifies DNA, to search for sequences from the Y chromosome, which could come only from the fetus. This method correctly flagged most of the pregnancies with male fetuses; conversely, it produced no signal from female fetuses. Furthermore, it required only a tiny quantity—10 microliters—of plasma or serum. Lo thus achieved a momentous step toward his goal. By identifying and analyzing cell-free fetal DNA in the blood of pregnant women, he opened a new avenue for developing non-invasive prenatal diagnostic tests.
To develop them, Lo needed to know how much fetal DNA was present—and how its quantities change during pregnancy. For this task, he deployed a PCR-based method that measures DNA after every round of copying rather than just at the end, and thus offers quantification rather than a plus/minus result. The average percentage fetal DNA concentration in plasma was significant; 3% early in pregnancy, 6% later. The absolute amount of fetal DNA in serum was similar as that in plasma, but a high background of maternal DNA made this material less attractive to work with. Lo could detect fetal DNA in the mother's plasma as early as the 7th week of gestation.
Other researchers had reported that fetal cells can remain inside the mother for up to 27 years. If DNA, too, boasted such endurance, it would complicate prenatal diagnoses, as analyses would be plagued by ghosts of former pregnancies. Fortunately, Lo found that nothing persisted in the mother's blood after two hours. This rapid clearance rendered his approach suitable for finding out about current rather than past fetuses.
Quantifying DNA
Lo wanted to implement the method in a situation where the results would guide medical treatment, and Rh factor status presented an opportunity. An Rh-negative woman, who does not make the Rh protein, can carry a baby who has inherited the Rh gene from its father. This situation is not problematic at that time, but exposure to the protein in subsequent pregnancies with Rh-positive fetuses can stir an antibody attack on the fetus. Treatment is available, but obstetricians need to know who to treat.
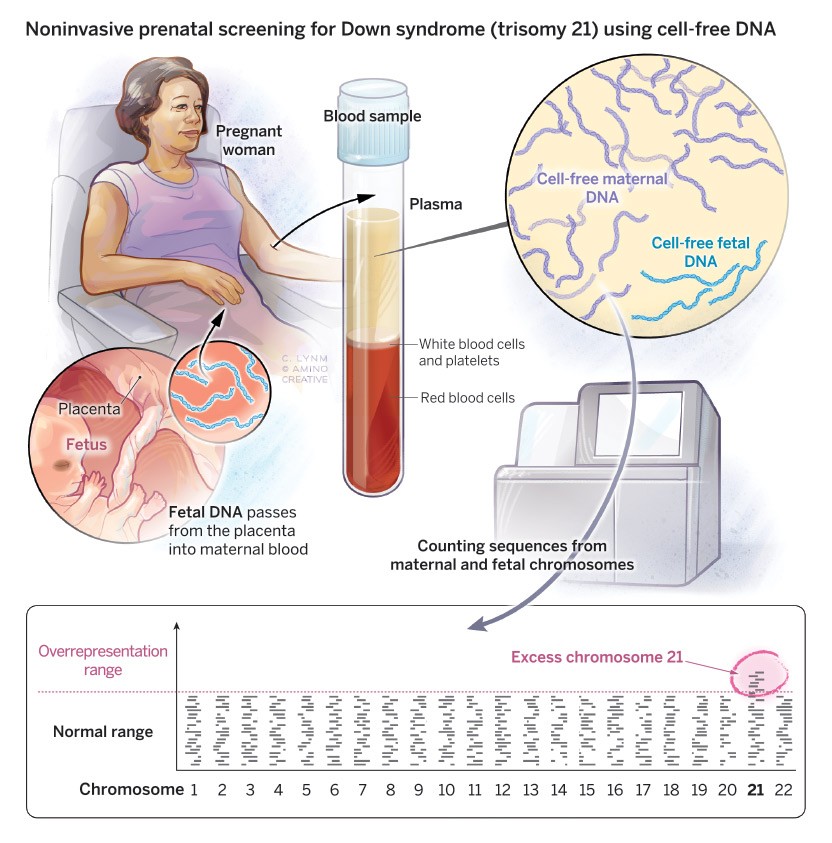
Counting chromosomes
Non-invasive prenatal testing exploits the presence of fetal genetic material in a pregnant woman's blood. A simple blood draw produces a plasma sample, which contains a mix of maternal and fetal DNA. Although the fetal material contributes only about 10% of the total, the technique is sensitive enough to detect small proportional changes in chromosome quantities. After amplifying random stretches of DNA with PCR, the pieces are sequenced and mapped to different chromosomes. In Down syndrome (trisomy 21), an excess of chromosome 21 appears.
Illustration: Cassio Lynm / © Amino Creative
In 1998, Lo examined fetal DNA in pregnant Rh-negative women. When deployed in the second or third trimester, his analysis correctly identified Rh status of 45 out of 45 babies eventually born to these women. These results suggested that the method could diagnose other conditions that arise from single-gene defects—cystic fibrosis and β-thalassemia, for instance—in partnerships where the father and mother harbor different and distinguishable versions of the culprit genes.
This progress heartened Lo, and he wondered whether he could harness the technology to reach the big prize in prenatal diagnosis: Down syndrome. To accomplish this feat, he would have to engineer a significant leap. Thus far, his strategies made use of genetic markers in the fetus that were absent in the pregnant woman. Now he would have to discern quantitative differences in the genetic marker—a chromosome—in a situation where it appeared in both of their bodies, but in distinctive proportions.
Lo devised a scheme that depended on the extra chromosome 21 carried by a trisomic fetus. This added dose would translate into an increased ratio of chromosome 21 relative to any other chromosome. The rationale was robust, but its execution presented challenges. He needed to find the excess chromosome 21 in a setting where the fetal DNA contributes a small fraction of the total. Based on the best measurement at that time, at least 10% of DNA in his plasma samples came from the fetus, but that DNA was still swimming in a sea of maternal material.
Years earlier, others had developed a method called digital PCR, in which researchers aliquot extremely dilute samples into tiny wells such that many wind up empty and most that contain DNA hold only one molecule. With this system, a positive signal after PCR amplification reflects a single original nucleic acid molecule and thus enables researchers to count the number of DNA molecules in a sample. By amplifying a chosen gene on chromosome 21 and a different gene on another chromosome, Lo could tally the proportion of chromosome 21 relative to a reference chromosome. Overrepresentation of the chromosome 21 gene sequence indicated trisomy 21, and Lo published this approach for detecting Down syndrome in 2007. Around the same time, Stephen R. Quake (Stanford University) reported similar results.
Although digital PCR provided an important step, it amplified only a single target gene on chromosome 21 and on the reference chromosome. If Lo could copy many distinct pieces of each chromosome, he could harvest much more information from the same quantity of plasma, thereby counteracting, to some degree, the huge dilution factor from maternal DNA.
Independently, Lo and Quake sequenced a multitude of random DNA fragments and then assigned them to their chromosome of origin, using the known human genome sequence. In 2008, both researchers demonstrated that this so-called massively parallel sequencing (MPS) strategy could rapidly and robustly count molecules. In Lo's report, 28 first and second trimester maternal plasma samples were tested. He identified all 14 trisomy 21 fetuses and all 14 normal fetuses.
Birth of a clinical test
Over the next several years, Lo deployed this method to validate the performance and feasibility of MPS for prenatal testing of Down syndrome. The first large-scale clinical study of the tactic reported on 753 pregnant women who were at high risk for trisomy 21 based on conventional screening. Lo and colleagues compared the results from his non-invasive approach with those from amniocentesis or chorionic-villus sampling. His technique identified all 86 fetuses with trisomy 21 and all 143 fetuses without trisomy 21; it incorrectly assigned trisomy 21 status to 3 of 146 fetuses without this condition. Reliance on these tests to rule out trisomy 21 would not have missed a single case and would have protected 98% of the women who were not carrying fetuses with Down syndrome and thus unnecessarily underwent the invasive procedures. Soon afterward, Lo reported encouraging results for trisomy 13 and trisomy 18.
This work culminated in the commercial launch of tests for Down syndrome in 2011. Centers around the world quickly embraced the technology and professional bodies endorsed them. More than 60 countries have adopted the tests, and in 2018, 10 million were performed. They are now used primarily to screen for trisomy 21, trisomy 18, trisomy 13, and sex chromosome imbalances.
The ability to measure amounts—rather than just the presence or absence—of relevant sequences offered possibilities beyond trisomy detection. For instance, it can allow scientists to determine whether a fetus has inherited one or two copies of a disease-causing mutation. This information is useful because some disorders manifest only if an individual carries a pair of faulty genes.
Lo has continued to hone and innovate, facilitating an ever more detailed view of the fetal genome. In a pioneering 2010 study, for instance, he mapped every variation in the genome to the mother or father through maternal blood, thus enabling scientists to spot multiple disorders as well as genetic alterations that arise during sperm and egg production.
Lo and others are also probing ways to exploit cell-free DNA in cancer management—to screen for disease and monitor therapy effectiveness—and in transplantation biology. Because DNA leaks when tissue dies, DNA from a donated organ can indicate rejection of the new tissue. He has not only made prenatal testing safer for millions of women and their fetuses, he has also spearheaded myriad other breakthroughs and his work has laid the groundwork for many more.
by Evelyn Strauss
Selected Publications – Discovery of Cell-free Fetal DNA
Lo, Y.M.D., Corbetta, N., Chamberlain, P.F., Rai, V., Sargent, H., Redman, C.W.G., and Wainscoat, J.S. (1997). Presence of fetal DNA in maternal plasma and serum. Lancet. 350, 485-487.
Lo, Y.M.D., Tein, M.S.C., Lau, T.K., Haines, C.J., Leung, T.N., Poon, P.M.K., Wainscoat J.S., Johnson, P.J., Chang, A.M.Z., and Hjelm, N.M. (1998). Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 62, 768-775.
Lo, Y.M.D., Lun, F.M.F., Chan, K.C.A., Tsui, N.B.Y., Chong, K.C., Lau, T.K.. Leung, T.Y., Zee, B.C.Y., Cantor, C.R., and Chiu, R.W.K. (2007). Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Natl. Acad. Sci. USA. 104, 13116-13121.
Fan, H.C., and Quake, S.R. (2007). Detection of aneuploidy with digital polymerase chain reaction. Anal. Chem. 79, 7576-7579.
Fan, H.C., Blumenfeld, Y.J., Chitkara, U., Hudgins, L., and Quake, S.R. (2008). Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA. 105, 16266-16271.
Tsui, N.B., Lun, F.M., Zee, B.C., Lau, T.K., Cantor, C.R., and Lo, Y.M.D. (2008). Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 105, 20458-20463.
Lo, Y.M.D. … Cantor, C.R., and Chiu, R.W. (2010). Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 1-13, 61ra91. doi: 10.1126/scitranslmed.3001720.
Chiu, R.W.K. … Lo, Y.M.D. (2011). Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. Br. Med. J. 342, 1-9, c7401. doi: 10.1136/bmj.c7401.
Chen, E.Z. … Lo, Y.M.D. (2011). Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS ONE. 6, 1-7, e21791. doi: 10.1371/journal.pone.0021791.
Ravitsky, V., Roy, M.-C., Haidar, H., Henneman, L., Marshall, J., Newson, A.J., Ngan, O.M.Y., and Nov-Klaiman, T. (2021). The emergence and global spread of noninvasive prenatal testing. Annu. Rev. Genom. Hum. Genet. 22, 309-338.




