Victor R. Ambros
University of Massachusetts Medical School

David C. Baulcombe
University of Cambridge

Gary B. Ruvkun
Harvard Medical School, Massachusetts General Hospital
For discoveries that revealed an unanticipated world of tiny RNAs that regulate gene function in plants and animals.
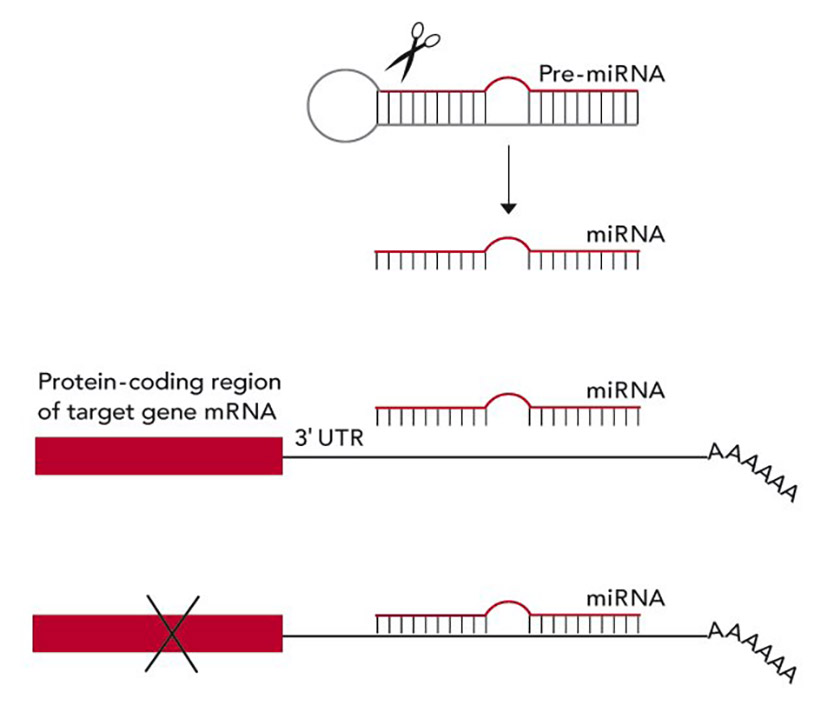
The 2008 Albert Lasker Award for Basic Medical Research honors three scientists who discovered an unanticipated world of tiny RNAs that regulate gene activity in plants and animals. Victor R. Ambros (University of Massachusetts Medical School, Worcester) and Gary B. Ruvkun (Massachusetts General Hospital, Boston, Harvard Medical School) unearthed the first example of this type of molecule in animals and demonstrated how the RNAs turn off genes whose activities are crucial for development. David C. Baulcombe (University of Cambridge) established that small RNAs silence genes in plants as well, thus catalyzing discoveries of many such RNAs in a wide range of living things. His findings led to the identification of the biochemical machinery that unifies numerous processes by which small RNAs govern gene activity.
Ambros, Baulcombe, and Ruvkun did not set out to unveil small regulatory RNAs. Ambros and Ruvkun were studying how the worm Caenorhabditis elegans develops from a newly hatched larva into an adult. Baulcombe, in a seemingly unrelated line of inquiry, was probing how plants defend themselves against viruses. All three investigators possessed the open mindedness, wisdom, and experimental finesse to entertain the possibility — and then verify — that tiny RNAs could perform momentous feats. Their work has led to the realization that these molecules are pivotal regulators of normal physiology as well as disease.
Award presentation by H. Robert Horvitz
The cell is the fundamental unit of life. Our bodies are made up of cells, about 10 trillion cells, and these cells are of many types: skin cells, blood cells, muscle cells, nerve cells, and so on. These cell types are strikingly different from each other, yet the genes they carry are, in almost all cases, identical. How can this be? The answer is that the expression of genes is controlled, so that some genes are expressed in some cells and other genes are expressed in other cells. The consequent difference in gene expression is what makes cells differ. Abnormalities in gene expression are responsible for many diseases, for example, many cancers. How gene expression is controlled is one of the fundamental and most important problems in biology.
Acceptance remarks

Acceptance remarks, 2008 Lasker Awards Ceremony
On my application for admission to MIT, my essay on the topic of “Why I want to attend MIT,” consisted of just six words: “I want to be a scientist.” My path, from that state of extreme naiveté in 1970 to this occasion today, would be impossible without an enormous web of support, consisting of my family, friends, teachers, mentors, bosses, colleagues, collaborators, and my wife Candy Lee. When I applied to MIT, I knew that if I hoped to become a scientist, I needed to be in the right place, but I did not know then that it would matter so much that I should be among the right people. This award is really in recognition of a confluence of events and people in my life, and it is my enormous pleasure that so many of those who have helped me are here today. In the spirit of enforced brevity, I will just say that you know who you are, and I refer everyone to my written comments for more details (and I should also mention a newcomer to my support net, Mike Czech, who hired me into my latest and best job, and who has become my friend and mentor).
I think my six-word MIT admissions essay was an attempt to distill a poorly defined adolescent dream — of belonging one day to a grand tradition populated by mythic figures: Einstein, Hubble, Galileo. But, what I learned at MIT, to my relief and delight, was that science is actually done by regular folks! To this day, what I love most about science is that it is such a deeply, intensely human enterprise. The success of the enterprise of science, and of the individual scientist, is derived precisely from the fact that we do it together. We work together in synergy as small teams, such as Candy Lee and Rhonda Feinbaum did to discover the first microRNA; we communicate our treasured, secret data across lab borders, as Gary Ruvkun and I did to enable us to discover the antisense base-pairing between microRNAs and targets; and we publish to the world our findings, so that for example, I opened a journal in 1999, and was astonished by a report from David Baulcombe’s group of little RNAs, just like lin-4, way over there in plants.
Not only do we do science as individuals, teams, and as a community of scientists, perhaps most importantly, science is an enterprise of and by the public. One does not have to be a scientist to contribute substantively to scientific discovery. We can be certain that without financial support from foundations and public agencies, particularly the National Institutes of Health, and without the commitment of universities to basic research, we would not be here today, and nobody would know anything about small RNAs, or about so many other things. For me, the Lasker Award is a profound personal honor, because I know that it recognizes not just the creative effort of Candy and Rhonda, Gary and his group, David and his co-workers, but it also emphasizes once again the vital legacy of Mary Lasker and her pioneering work on behalf of publicly supported science in our modern era. Science is among the best things that we do as a species, and I am proud and happy beyond words to be part of it.

Acceptance remarks, 2008 Lasker Awards Ceremony
My initial reactions to hearing about the Lasker award were surprise, delight, satisfaction, gratitude to colleagues, and a nice warm feeling. I was also pleased that there had been recognition of plants as model systems — I have written about that in the commentary article. After a while, I also started wondering how I got into this prize-winning position. Part of the answer of course is luck and having great colleagues. The other part is because EVENTUALLY we — my group — did what we should do as scientists. Through a process of deduction and controlled experimentation we worked out that the important molecules are antisense of the target molecule and they are small and finally we made some progress.
Now the point I would like to make is about the word “EVENTUALLY” — the whole process took some considerable time, and it was a while before we even realized that we were looking at something extraordinary. Even then, we messed about with misguided models based on proteins or even DNA interactions before we got on the right track. What we did initially was what all people do most of the time — we were jumping to conclusions. The right side of our brain took over and our thinking was based on preconceptions, hunches, and incomplete information.
In everyday life, the intuitive right hemisphere process serves us quite well: it would be a pretty strange and feeble existence without it. Right hemisphere thinking may even have a proper place in the early pathway of a scientific discovery. But, for most of the time in science, the evidence-based logical left hemisphere should dominate.
Now, the point of all of this is that practically everyone on the planet has a functioning left hemisphere, and potentially everyone could embrace the scientific way of thinking from time to time, but they do not. Perhaps if people could understand that many scientists struggle with this right and left hemisphere conflict, it would be easier for them to accept science and what it has to offer. I see getting widespread acceptance of science and its technological opportunities as being a significant challenge — even now. The Lasker Foundation has established an excellent forum to promote science and I am honored to be associated with it.

Acceptance remarks, 2008 Lasker Awards Ceremony
The discovery of a previously unsuspected civilization of tiny RNAs was the first perfect storm of the genome era. I would like to acknowledge the inspired discoveries of my co-awardees Victor Ambros and David Baulcombe, whom I have now known for most my scientific career. It is important for those interested in medical research to realize that much of the tiny RNA revolution emerged from non-mammalian genetic analysis — from plant biology, from worm genetics, from the fungi, from the protozoa. Much of this research was funded generously over the past decade or two, reflecting well on the wisdom of the many peer review systems, private and public, which support science. It is important to continue to explore the diversity of biology; our next revolutions, scores of them, are likely to emerge from here.
It was Sputnik in 1957 that launched me into science. The ensuing US launches were fully televised and breathlessly reported. Television was a new medium, and unlike today, with the inevitable launch delays, there could be four or five hours of continuous network coverage, with hours of prerecorded interviews with bowtied, flattop haircut scientists to fill the time. I fondly remember the excitement of Walter Cronkite and Jules Bergman when they interviewed the engineers who designed space capsules or the scientists who described orbital mechanics. The exciting concepts that I learned from television those few dozen mornings from 1961 to 1966 were far more sophisticated than what was being taught in my elementary school and were key elements in shaping me.
I thank my parents for my first microscope at age 5, for my telescope at age 8, for taking me to the Oakland Public Library once a week to check out a dozen books, for teaching me how a steel mill works, for my first ham radio, for their celebration of Berkeley as Athens, for the awe in their voices when they said, “nuclear physicist.”
One of the most wonderful dividends of this sort of recognition is that I can interpret aspects of my autobiography, exploring how my past might have helped me come to discoveries. And that there are audiences that I can try to interest in my saga. It is at least possible that being a Jew in Oakland, California, made me enough of an outsider that I did not neatly fit into any community and escaped a sense of entitlement. And that my kinship with the great Jewish comics of the ’50s and ’60s fostered a certain irreverance and a warped perspective,and made me funny so that people might gravitate to work with me. It is possible that my undergraduate training in physics taught me not quantitative reasoning but rather how a scientific revolution at its very beginning can be recognized. And that my year working in a tree-planting cooperative in Oregon after graduating from college taught me the joys of a crew working together, cooperatively, on a hard problem. And that my year of travel on third-class buses from Mexico to Tierra del Fuego, with my encounters with thousands of strangers, may have trained me in the instantaneous assessment of the others, no doubt helping me to recruit the most interesting and intelligent students and post-docs to my lab.
I arrived at graduate school in 1976, with no idea how science is done. From my fellow graduate students and post-docs at Harvard and MIT, I learned how to eat and breathe science, how to be thrilled with discoveries at the moment of their presentation. From my teachers, Fred Ausubel, Wally Gilbert, and Bob Horvitz, all of whom are here today, I learned something more intangible but key: good taste in picking scientific problems, and by their examples, how to unravel the tangled webs that life has evolved.
So the tribes of my education, my childhood, my lab, and my academic environment were uniquely inspiring, supportive, and loads of fun. But my home tribe has been the wellspring of strength and joy. And I am truly grateful to Natasha Staller, and our daughter Victoria, for the joyous home life of our little tribe.
Interview with Victor R. Ambros, David C. Baulcombe, and Gary B. Ruvkun
Video Credit: Susan Hadary