Too simple, too uniform. Those were the arguments that scientists made for nearly a century to explain why DNA was very likely not the blueprint for life that we consider it to be today.

DNA in water [Courtesy of Bbkkk, CC BY-SA 4.0, via Wikimedia Commons]
But experiments in the 1940s and 1950s made DNA a prime candidate for hereditary material. The finding by Alfred Hershey and Martha Chase in 1952 that it is the DNA, not the protein, of bacteriophages that gets inserted into bacteria during infection made the most convincing case at the time that genes are actually made of DNA. “That’s when the race began to find the structure of DNA,” says Evelyn Witkin, a 2015 Lasker Laureate. The moment that James Watson and Francis Crick described the now-iconic double helix in their famous 1953 publication, there was no looking back. “It was like a curtain being lifted and you could just see how it worked,” Witkin says of the meeting at Cold Spring Harbor, where she was based at the time, at which she first saw the DNA double helix model. “That was a high point that is hard to duplicate,” she adds.
In the span of just a couple of years the door was thrown open, after decades of being sealed shut, to the paramount importance of DNA. Its structure explained, almost intuitively, how it could be replicated as cells divide and how its information could be transmitted during reproduction. Although it was still formally only a “working hypothesis” that genes were made of DNA, it was getting harder to deny.

Walter Gilbert
And yet, the model of DNA explained little about its functions in the cell. One question rose to the fore, as Lasker Laureate Walter Gilbert put it:
“How does DNA do anything?”
As the following decades revealed, what DNA does — how it is read, regulated, and repaired — turned out to be deceptively complex for such a seemingly simple molecule. From the breakneck progress cracking the genetic code to controversies over editing genomes, Lasker Laureates led some of the innumerable advances over the past 60 years. No one appreciates more than they do the central role that DNA plays in understanding life. “Historically and biologically, our understanding of DNA has been the driving force in all of this,” says Tom Maniatis, recipient of a 2012 Lasker Award.
Learning the Language of DNA
As soon as DNA was cast in the role of likely carrier of genetic information, a frenzy was unleashed in the scientific world to decipher the language of DNA’s four nucleotides. How could the As, Ts, Gs, and Cs, in sets of different sequences and lengths, spell out genes? And how could these genes convey meaning that would get passed on to daughter cells and daughter organisms?

Meeting of the members of the RNA Tie Club, Cambridge, UK Fall 1955 (from upper left): Francis Crick, Leslie Orgel, Alexander Rich and James D. Watson [Courtesy of the James D. Watson Collection, Cold Spring Harbor Laboratory Archives]
Nevertheless, it was discussions within the club that inspired Crick to propose at a symposium in London in 1957 that the main function of DNA is to make proteins and that RNA, which had long been suspected to be important for protein synthesis, is some kind of intermediary between DNA and protein.
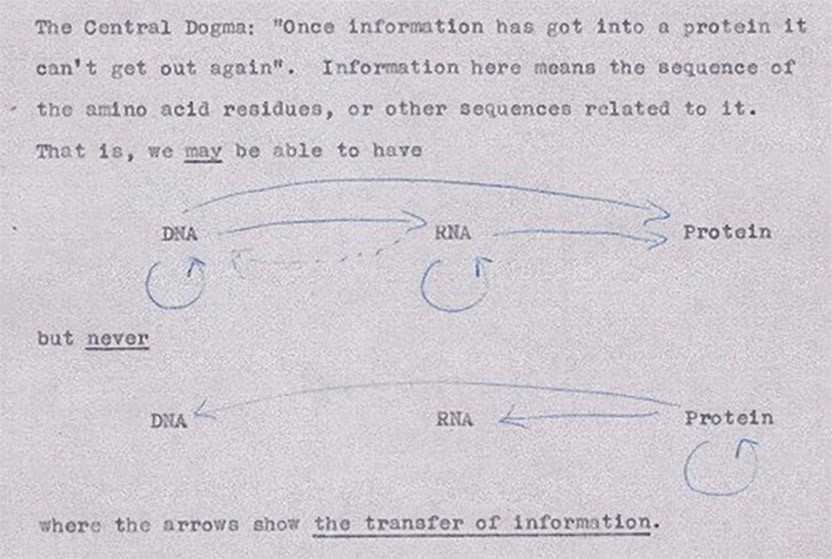
Crick’s first illustration of the central dogma, from an unpublished note in 1956
The proposal may have been jarring at the time, but it is now the central dogma of biology. In keeping with this proposal, the gentleman’s club, which was dubbed the RNA Tie Club, assigned each of the 20 members one of the 20 amino acids with the mission to determine the nucleotide combinations that encode it.
In one of the biggest advances since discovering the structure of DNA, Francis Crick and Sydney Brenner, a fellow RNA Tie Club member and recipient of two Lasker Awards, led work at the University of Cambridge, UK that mapped out how nucleotides are read. Their experiments revealed that the magic number of nucleotides encoding a single amino acid was three, and nucleotide triplets, or codons, did not overlap.

Marshall Nirenberg in the 1960s [Courtesy of National Institutes of Health]
Cracking the genetic code, and learning how the linear molecule of DNA determines the three-dimensional structure of proteins, marked the dawn of molecular biology. “It was a lovely conception and idea behind molecular biology,” recalls Lasker Laureate Gilbert, who was so enthralled by the burgeoning field that he abandoned his physics studies to do molecular biology research.
In the watershed period of the early 1960s, there was also intense focus on describing the mysterious transient RNA molecule — which was dubbed “messenger RNA” or mRNA — that was thought to carry the information in DNA to ribosomes in the cytoplasm to instruct protein synthesis. Although identifying mRNA ultimately involved numerous scientists and multiple experimental approaches, Gilbert recalls many details of the experiments done by his team in Watson’s lab at Harvard University, including pouring radioactive phosphate into a bacterial culture and purifying the molecule from cells. It was becoming clear, in leaps and bounds, how the three main players — DNA, RNA, and protein — worked together in the machine of the cell to read the language of genes.
Revealing Gene Regulation
As the early puzzles were solved about the genetic code and how the information stored in DNA is converted into proteins, scientists were flocking to more nuanced questions about gene regulation. What controls when genes are expressed and their levels of expression?
“The next hallmark in the whole progression of molecular biology… [came from] Francois Jacob and Jacque Monod,” who gave the first hints about how genes are controlled, says Phillip Sharp, 1988 Lasker Laureate. The pair of scientists at the Pasteur Institute, who were also instrumental in describing mRNA, found a gene that repressed gene expression. But the French scientists had no idea how the product of this gene carried out this activity and suspected it was through an RNA molecule. Then in 1966, Gilbert, along with Mark Ptashne also at Harvard, found that repressors are proteins that bind to DNA.

Tom Maniatis at Cold Spring Harbor Laboratory in 1977 [Courtesy of the Cold Spring Harbor Laboratory Archives]
By the 1970s, research on gene regulation had become so competitive that some scientists avoided it altogether. “Everybody and their brother or sister were studying how RNA polymerase interacts with [regulatory] factors to transcribe a gene. What I wanted to do was work on something that was not crowded,” recalls Michael Grunstein. When he started his own research group in the mid-1970s, Grunstein decided to study “boring” histone proteins and how they package DNA into chromosomes. The plan yielded dramatic and unexpectedly exciting results when he started to notice that yeast mutants he engineered to lack one type of histone protein had higher levels of expression of certain genes, suggesting that seemingly inert histones might actually repress transcription in the living cell. Moreover, sites of posttranslational modification (acetylation) in histones were shown to be required for gene activity in vivo. Thus, his work clarified that histones and their acetylation sites help regulate transcription. Several years later, C. Davis Allis, who shares a 2018 Lasker Award with Grunstein, identified the enzyme that adds chemical groups to certain amino acids in histones and turns on gene expression.
Eager to push the understanding of gene regulation even further, scientists moved into new technology frontiers: DNA sequencing and gene cloning. As Maniatis says, it was becoming clear that “one could not even imagine studying gene expression in any detail in eukaryotic cells without recombinant DNA,” whose advent was right around the corner.
Taming a Messy Molecule
Just as scientists did not grasp the function of DNA until well after they appreciated the roles of RNA and proteins, their ability to sequence DNA trailed behind their ability to do so for RNA and proteins. In fact, most scientists thought that DNA sequencing was not possible, but the research teams of Gilbert and Frederick Sanger at the University of Cambridge (both recipients of a 1979 Lasker Award and a 1980 Nobel Prize) persevered. “Various members of the [Sanger] lab were all trying different methods for sequencing DNA,” recalls Elizabeth Blackburn, winner of a 2006 Lasker Award, of her PhD with Sanger in the early 1970s. In fact, one of the early success stories involved copying a stretch of DNA into RNA and sequencing the RNA fragments. Using that approach in 1974, it took Gilbert about two years to read 20 nucleotides in the bacterial repressor binding site.

Fred Sanger at Laboratory of Molecular Biology, UK, circa 1969 [Courtesy of MRC Laboratory of Molecular Biology]
The 1970s saw turning points in the ability not just to sequence DNA but also to study and manipulate it. Donald Brown, recipient of a 2012 Lasker Award, recalls trying to work with DNA in the late 1960s, and it “was just a glop, a mess.” But certain DNA fragments, such as the ribosomal RNA (rRNA) genes that Brown and 2006 Lasker Award recipient Joseph Gall worked with, had properties that made their isolation possible even before the days of recombinant DNA. With the isolated rRNA genes in hand, Blackburn was able to sequence the telomeres at their ends. The rRNA genes also played a pivotal part in the early days of recombinant DNA. Brown sent the purified genes to Herbert Boyer and Stanley Cohen, two of the four scientists to receive a 1980 Lasker Award for developing recombinant DNA tools such as restriction enzymes and bacterial plasmids. In a now-classic experiment, Boyer and Cohen inserted fragments of the genes into a plasmid that they then delivered into Escherichia coli and demonstrated that bacteria could make copies of foreign DNA. Brown proceeded to use these rRNA clones to uncover some of the first details about gene regulation in eukaryotes.

Elizabeth Blackburn in 1975 [Courtesy of Elizabeth Blackburn]
With the advent of modern molecular biology, concerns were growing that recombinant DNA might present risks, and certain research institutions declared a moratorium on such research. But some of the scientists who initially most vehemently opposed recombinant technology later became staunch supporters of the fledgling Human Genome Project, which by 2003 had relied on the technology to sequence entire human genomes.
Scientists have used sequencing and recombinant DNA technologies to determine the function of genes, how genes make us who we are, and contribute to human disease. In the late 1970s, scientists gained additional tools: Southern blotting and DNA fingerprinting, developed by 2005 Lasker Award recipients Edwin Southern and Alec Jeffreys, respectively. Previously, to scan for specific DNA sequences within genomes, scientists had to grapple with a long, diffuse smear of genomic fragments in an agarose gel, after separating the fragments using gel electrophoresis. Southern devised a technique to transfer the DNA smear to a membrane that could be easily probed using radioactive complementary RNA fragments, and with it, scientists quickly identified sequence differences in genes associated with sickle cell anemia and other diseases. Alec Jeffreys made use of Southern blotting to find regions across the human genome that are highly variable, and he determined how to probe for a combination of such regions to distinguish unique human genomes. This DNA fingerprinting technique was first used in the 1980s to verify a person’s identity and continues to be central to forensic science.
Another major leap forward in understanding human disease was the advent of knockout mice.

Mario Capecchi in the lab in the 1980s [Courtesy of Mario Capecchi]
Looking Ahead to the Next Advances
As early as the 1960s, before some of the greatest breakthroughs in molecular biology were even on the horizon, scientists were saying the field was dead. Christiane Nusslein-Volhard, recipient of a 1991 Lasker Award, recalls hearing that prediction when she was working toward her PhD in the early 1970s.
But as it turned out, of course, molecular biology provided the scientific community with both groundbreaking insights and puzzles so confounding that they are still being worked out. Even the most fundamental biological processes, such as gene transcription, are not fully understood, says Stephen Elledge, recipient of a 2015 Lasker Award. Many goals of molecular biology, such as identifying associations between DNA sequence variants and disease, remain the same but have expanded into more complex systems, such as neuropsychiatric diseases, Maniatis says. In some cases, as researchers delve deeper into one aspect of molecular biology, they find connections with other areas. In her ongoing quest to elucidate how telomere length is controlled, Carol Greider, recipient of a 2006 Lasker Award, has ventured into research on DNA replication and how that process is linked to telomere regulation.
DNA molecule [Courtesy Zephyris, CC BY-SA 3.0, via Wikimedia Commons]
By Carina Storrs
